Gene Mutation Prediction using Deep Learning
Multiple Instance Dcision Aggregation for Gene Mutation Detection from CT
Kidney clear cell renal cell carcinoma (ccRCC) is the major sub-type of RCC, constituting one the most common cancers worldwide accounting for a steadily increasing mortality rate with 350,000 new cases recorded in 2012. Understanding the underlying genetic mutations in ccRCC provides crucial information enabling malignancy staging and patient survival estimation thus plays a vital role in accurate ccRCC diagnosis, prognosis, treatment planning, and response assessment. Recent studies have identified several mutations in genes associated with ccRCC. For example, the von Hippel-Lindau (VHL) tumor suppressor gene, BRCA1-associated protein 1 (BAP1) gene, polybromo 1 (PBRM1) gene, and SET domain containing 2 (SETD2) gene have been identified as the most commonly mutated genes in ccRCC. Although these underlying gene mutations can be identified by whole genome sequencing of the ccRCC following invasive nephrectomy or kidney biopsy procedures, recent studies have suggested that such mutations may be noninvasively identified by studying image features of the ccRCC from Computed Tomography (CT) data. Such image feature identification currently relies on laborious manual processes based on visual inspection of 2D image slices that are time-consuming and subjective. We proposed a convolutional neural network approach for automatic detection of underlying ccRCC gene mutations from 3D CT volumes. We aggregate the mutation-presence/absence decisions for all the ccRCC slices in a kidney into a robust singular decision that determines whether the interrogated kidney bears a specific mutation or not. When validated on clinical CT datasets of 267 patients from the TCIA database, our method detected gene mutations with 94% accuracy.
Multiple Instance Decision Aggregation for Mutation Detection

Typically, ccRCC grows in different regions of the kidney and is clinically scored on the basis of their CT slice-based image features, such as size, margin (well- or ill-defined), composition (solid or cystic), necrosis, growth pattern (endophytic or exophytic), calcification etc. We proposed to learn corresponding features from the CT images using four different CNNs: VHL-CNN, PBRM1-CNN, SETD2-CNN and BAP1-CNN, each for one of the four mutations (VHL, PBRM1, SETD2 and BAP1). Using a separate CNN per mutation alleviates the problem of data imbalance among mutation types, given that the mutations are not mutually exclusive. CNN Architecture: All the CNNs in this study (i.e. VHL-CNN, PBRM1-CNN, SETD2-CNN and BAP1-CNN) have similar configuration but are trained separately (Fig. 1). Each CNN has twelve layers excluding the input: five convolutional (Conv) layers; three fully connected (FC) layers; one softmax layer; one average pooling layer; and two thresholding layers. All but the last three layers contain trainable weights. The input is the 227×227×3 pixel image slice containing the kidney+ccRCC. We train these CNNs (layers 1–9) using a balanced dataset for each mutation case separately (i.e. a particular mutation-present and absent). During training, images are fed to the CNNs in a randomly shuffled single instance fashion. Typically, Conv layers are known for sequentially learning the high-level non-linear spatial image features (e.g. ccRCC size, orientation, edge variation, etc). We used five Conv layers as the 5th Conv layer typically grabs an entire object (e.g. ccRCC shape) in an image even if there is a significant pose variation. Subsequent FC layers prepare those features for optimal classification of an interrogated image. In our case, three FC layers are deployed to make the decision on the learned features from the 3-ch images to decide if a particular gene mutation is probable or not. The number of FC layers plays a vital role as the overall depth of the model is important for obtaining good performance, and we achieve optimal performance with three FC layers. Layers 10, 11 and 12 (i.e. two thresholding and one average pooling layers) of the CNNs are used during the testing phase and do not contain any trainable weights.
Data
We obtained access to 267 patients’ CT scans from The Cancer Imaging Archive (TCIA) database. In this dataset, 138 scans contained at least one mutated gene because of ccRCC. For example, 105 patients had VHL, 60 patients had PBRM1, 60 patients had SETD2, and 20 patients had BAP1 mutations. In addition, some of the patients had multiple types of mutations. However, 9 patients had CT scans acquired after nephrectomy and, therefore, those patients’ data were not usable for this study. The images in our database included variations in CT scanner models, contrast administration, field of view, and spatial resolution. The in-plane pixel size ranged from 0.29 to 1.87 mm and the slice thickness ranged from 1.5 to 7.5 mm. Ground truth mutation labels were collected from the cBioPortal for Cancer Genomics.
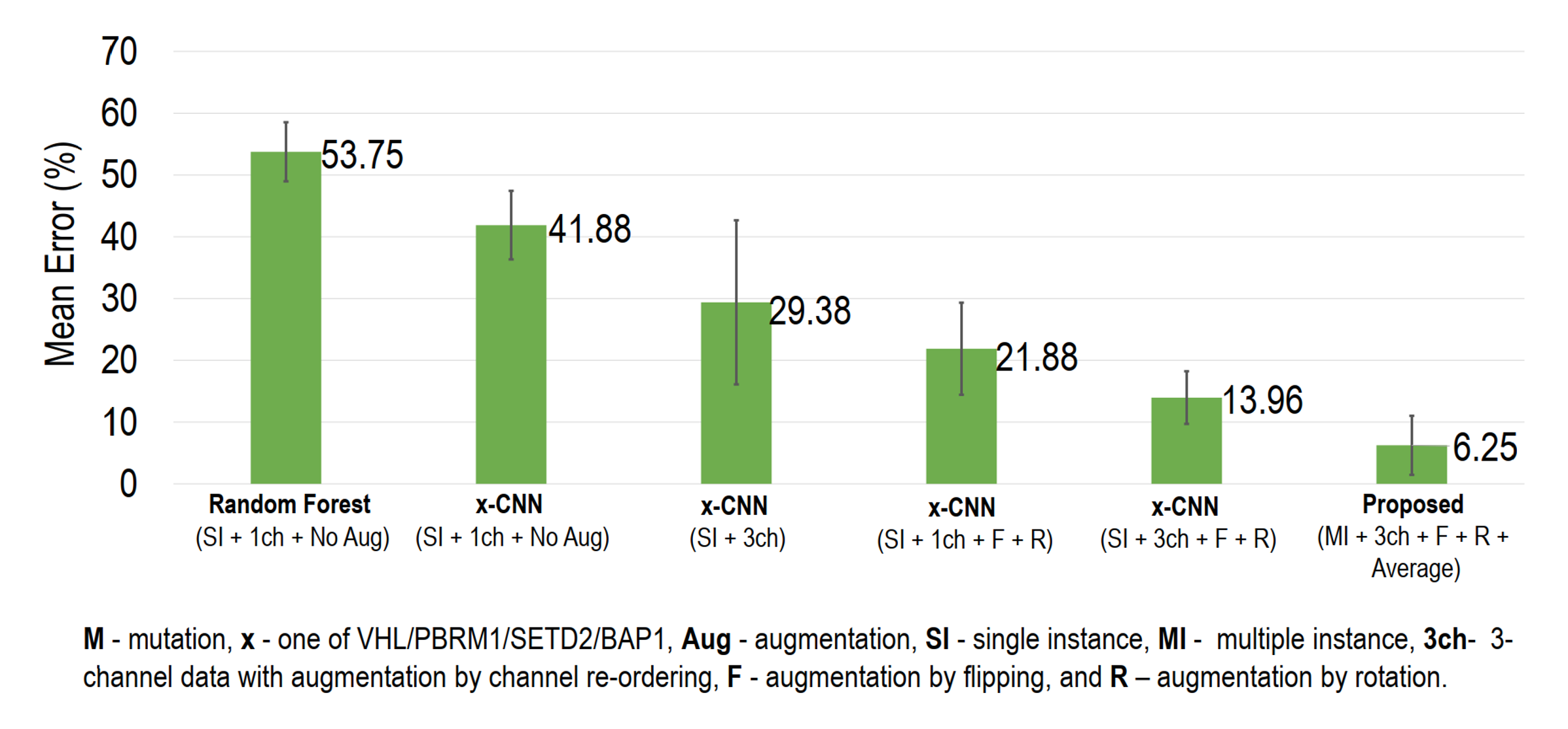
Results
For Details
Please read our [paper].