Kidney Assessment with Regression Forests
Segmentation-free Kidney Volume Estimation
The economic burden of chronic kidney disease (CKD) is significant, estimated in Canada in 2007 at $1.9 Billion just for patients with end-stage renal disease (ESRD). In 2011, about 620,000 patients in United States received treatment for ESRD either by receiving dialysis or by receiving kidney transplantation. ESRD is the final stage of different CKDs, e.g., Autosomal dominant polycystic kidney disease (ADPKD), renal artery atherosclerosis (RAS), which are associated with the change of kidney volume. However, detection of CKDs are complicated; multiple tests such as the estimated glomerular filtration rate (eGFR) and serum albumin-to-creatinine ratio may not detect early disease and may be poor at tracking progression of disease. Recent works have suggested kidney volume as the potential surrogate marker for renal function and is thus useful for predicting and tracking the progression of different CKDs. In fact, the total kidney volume has become the gold-standard image biomarker for the ADPKD and RAS progression at early stages of this disease. In addition, the renal volumetry has recently emerged as the most suitable alternative to renal scintigraphy in evaluating the split renal function in kidney donors as well as the best biomarker in follow-up evaluation of kidney transplants. Consequently, estimation of the ‘volume’ of a kidney has become the primary objective in various clinical analyses of kidney. Traditionally, the kidney volume is estimated by means of segmentation. However, existing kidney segmentation algorithms have various limitations (e.g., requiring user interaction, sensitivity to parameter setting, heavy computation). Therefore, in this project, we proposed a segmentation-free, supervised learning approach that addresses the challenges of accurate kidney volume estimation caused by extensive variations in kidney shape, size and orientation across subjects. We develop dual regression forests to simultaneously predict the kidney area per image slice, and kidney span per image volume.
Segmentation-Free Estimation of Kidney Volumes using Dual Regression Forests
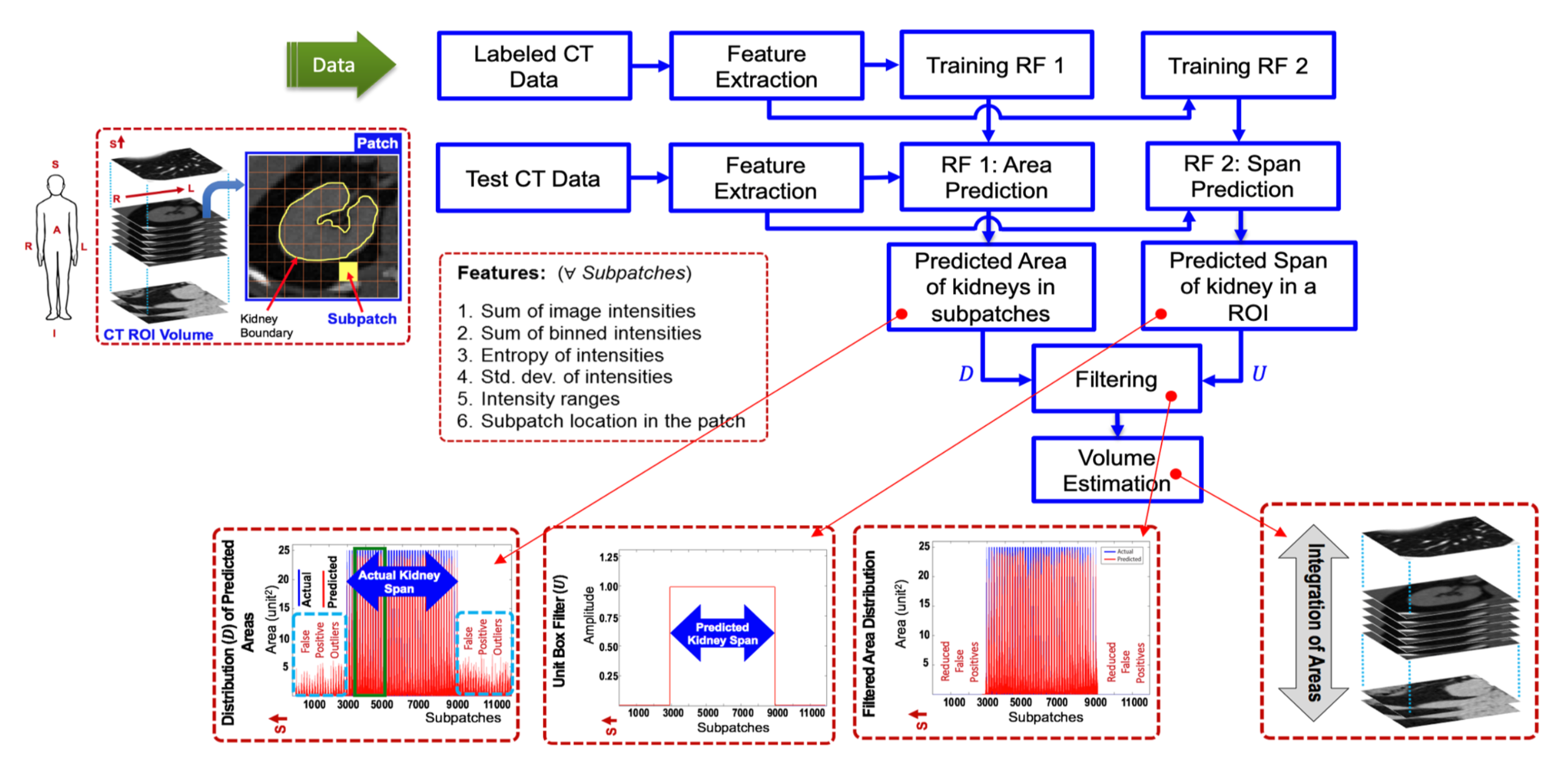
Regression forest 1 (Fig. 1) learns the correspondence between input features and kidney areas for training subpatches, and then predicts kidney areas in unseen subpatches. The distribution of estimated kidney area values vs. subpatches for a kidney sample is shown at the bottom in Fig. 1 (left most). However, due to extensive variation in kidney shapes, sizes and orientations across subjects, we observed that non-zero volumes are predicted for areas devoid of kidney tissue. These false positives are removed using a spatial filter (Fig. 1, second from the left at the bottom) having an extent (or bandwidth) equal to a spatial kidney span measure (along superior-inferior direction). This important span parameter is learnt by forest 2. For training forest 2, we use the same features. We define a unit step function whose spatial bandwidth is equivalent to the span predicted by forest 2 for a particular kidney sample. We approximate the most probable kidney span in the false positives-corrupted distribution by calculating the cross-correlation between distribution and span filter. Finally, we estimate the volume of a kidney by integrating the areas in the axial direction.
Data
We used 100 patients’ CT scans accessed from the Vancouver General Hospital (VGH) records with required ethics approvals by the UBC Clinical Research Ethics Board (CREB), certificate number: H15-00237. There were a total of 200 kidney samples, and we used 130 samples (from 65 randomly chosen patients) for training, 20 samples (from 10 randomly chosen patients) for validation, and the remaining 50 samples for testing. Our dataset included 12 pathological kidney samples (with endo- and exophytic tumors), and our training and test data contained six pathological cases each. We made sure that kidneys from the same patient were not split across the training, validation, and test cases. These data were acquired using a Siemens SOMATOM Definition Flash (Siemens Healthcare GmbH, Erlangen, Germany) CT scanner. Ground truth kidney bounding boxes were calculated from manual kidney delineation performed by an expert radiologist.
Results
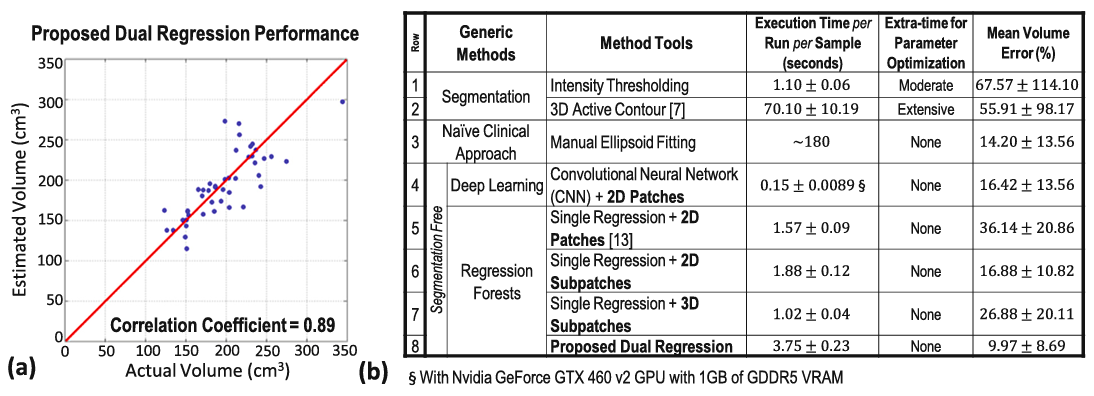
For Details
Please read our paper [1].