Forward Inclusion Backward Elimination
Efficient Feature Selection Using Forward Inclusion Backward Elimination for Machine Learning
This algorithm performs a feature selection for both regression and classification tasks using any of the following models (1) linear support vector regressor/classifier, (2) Gaussian support vector regressor/classifier, (3) Regression/random forest, (4) AdaBoost regressor/classifier with linear support vector and (5) AdaBoost regressor/classifier with decision trees. This algorithm can also use model consensus in feature selection using (1) linear support vector regressor/classifier, (2) Gaussian support vector regressor/classifier, and (3) Regression/random forest. For loss calculation as well as validation performance estimation, this algorithm comes with options of using (1) mean absolute error (MAE) (2) mean absolute percentage error (MAPE), (3) accuracy, (4) F1-score, and (5) binaryROC metrics. The fibe_function contains all the necessary functions to run this algorithm.
GitHub Repo
[https://github.com/i3-research/fibe]
How to Run the Algorithm
To run this algorithm, the following function is needed to call with appropriate parameter selection:
selectedFeatures, actualScore, predictedScore, validationPerformance = fibe(feature_df, score_df, fixed_features=None, columns_names=None, task_type=None, balance=False, model_name=None, metric=None, voting_strictness=None, nFold=None, maxIter=None, verbose=True)
Here,
-
feature_dfis the 2D feature matrix (supports DataFrame, Numpy Array, and List) with columns representing different features. -
score_dfis the 1D score vector as a column (supports DataFrame, Numpy Array, and List). -
fixed_featuresPredefined features that must stay in the feature set and the FIBE algorithm does not add or remove those. Must be either a List of names to select from ‘feature_df,’ or DataFrame of features added separately to ‘feature_df.’ -
columns_namescontain the names of the features. The algorithm returns the names of the selected features from this list. If not available, then the algorithm returns the column indexes of selected features. -
task_typeeither ‘regression’ or ‘classification.’ The default is ‘regression.’ -
balanceIn a binary classification task, if the data is imbalanced in terms of classes, ‘balance=True’ uses resampling to balance the data. -
model_nameFor ‘regression’ task, to choose from ‘linerSVR’, ‘gaussianSVR’, ‘RegressionForest’, ‘AdaBoostDT’, ‘AdaBoostSVR’, and ‘consensus’ (consensus using ‘linerSVR’, ‘gaussianSVR’, and ‘RegressionForest’). Default is'linerSVR'. For the ‘classification’ task, choose from ‘linerSVC’, ‘gaussianSVC’, ‘RandomForest’, ‘AdaBoostDT’, ‘AdaBoostSVC’, and ‘consensus’ (consensus using ‘linerSVC’, ‘gaussianSVC’, and ‘RandomForest’). The default is'linerSVC'. -
metricFor theregressiontask, choose from ‘MAE’ and ‘MAPE’. The default is ‘MAE.’ For the ‘classification’ task, choose from ‘Accuracy’, ‘F1-score’, and ‘binaryROC’. The default is'Accuracy'. -
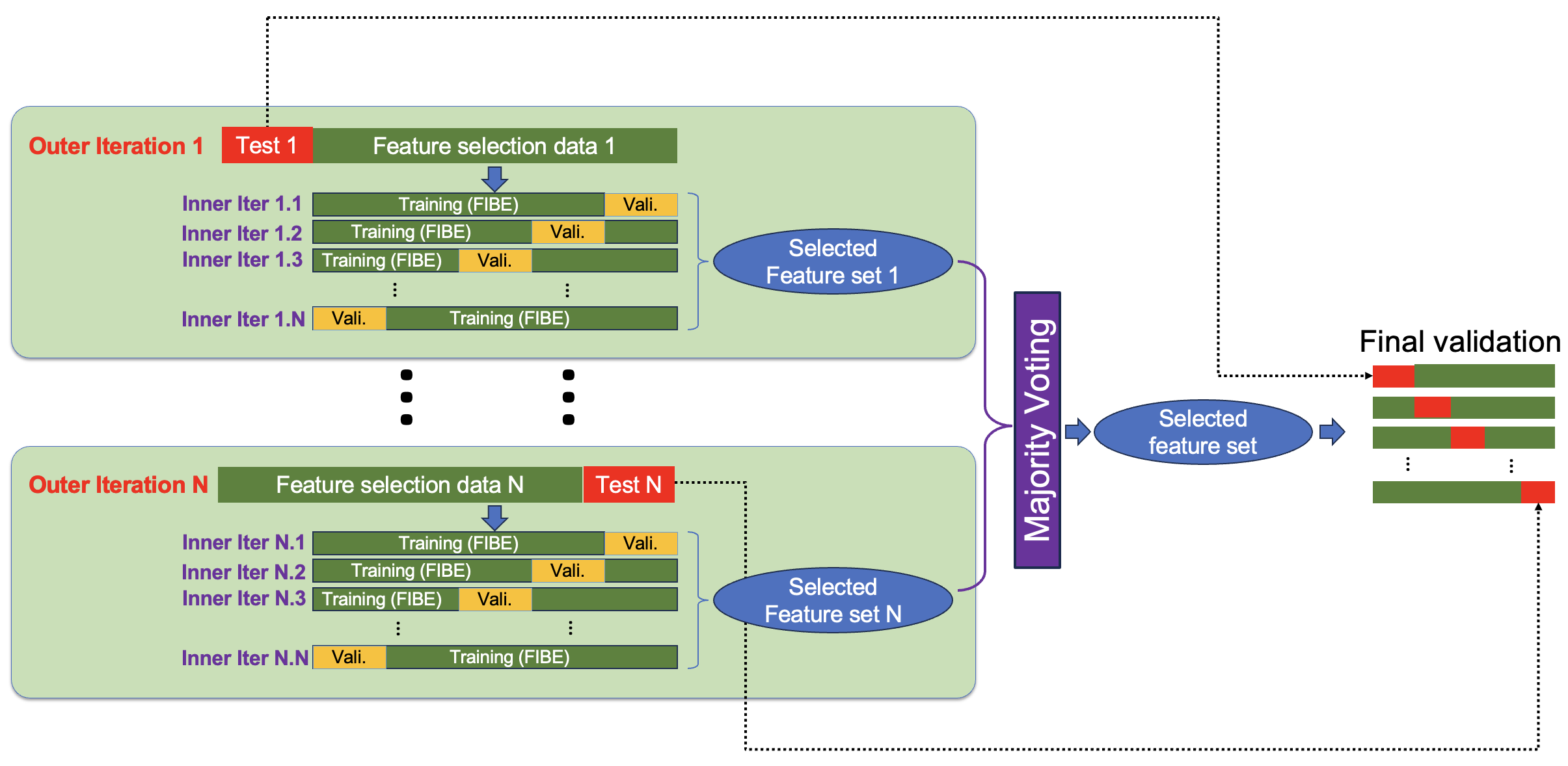
voting_strictnessThe option ‘strict’ chooses those features that are selected at least 3 times in 5-fold cross-validation; the option ‘loose’ chooses those features that are selected at least 2 times in 5-fold cross-validation, and the option ‘both’ produces two sets of results, one for ‘strict’ and one for ‘loose’. The default is'strict'. For any random number of folds,N, the ‘strict’ threshold should be0.6 X Nand the ‘loose’ threshold should be0.4 X N. -
nFoldNumber of folds in cross-validation. Preferred and default is5. -
masteris the maximum number of iterations that the algorithm goes back and forth in forward inclusion and backward elimination in each fold. The default is3. -
verbosegenerates text for intermediate loss and selected feature list during iteration. The default isTrue.
The outputs are:
-
selectedFeaturesis the list of features ifcolumns_nameswas notNone. Otherwise column indexes of the selected features. Forvoting_strictnessof ‘both’,selectedFeaturescontains two sets of output as[[selected features for 'strict'], [selected feature for 'loose']]. -
actualScoreis the list containing actual target scores. Ifmodel_nameis chosen as ‘consensus’, this list has a repetition of values 3 times, to correspond to predictions by three models. Forvoting_strictnessof ‘both’,actualScorecontains two sets of output as[[actual scores for 'strict'], [actual scores for 'loose']]. -
predictedScoreis the list containing predicted scores. Ifmodel_nameis chosen as ‘consensus’, this list has 3 predictions per observation. Although 3 predictions per observation are generated here, ‘consensus’ uses an averaging of the losses for 3 predictions in decision-making. Forvoting_strictnessof ‘both’,predictedScorecontains two sets of output as[[predicted scores for 'strict'], [predicted score for 'loose']]. -
validationPerformanceis a list containing validation performance in terms of chosenmetricfornFoldfolds. Forvoting_strictnessof ‘both’,validationPerformancecontains two sets of output as[[validation performance for 'strict'], [validation performance score for 'loose']].
Algorithm Overview
Example Code
An example Python file main.py is given. It includes example code to run one classification and one regression problem. Further, it includes examples of how to run the algorithm with predefined fixed features as well as data balancing options.