Segmentation with Semi-supervised Deep Learning
Lung and Infection Segmentation in COVID-19 CT
The coronavirus disease 2019, abbreviated as COVID-19, is currently a World Health Organization (WHO)-announced global pandemic. COVID-19 is caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). It is a highly contagious disease and although known to be transmitted primarily from human-to-human via respiratory fomites, a recent study also found evidence of live SARS-CoV-2 virus in human feces. COVID-19 is primarily known to affect the human respiratory system, however, it often increases the risk of death to patients with underlying cardiovascular disease. With the onset of the COVID-19 in December 2019, the polymerase chain reaction (PCR) assay of the sputum has been considered the gold standard for COVID-19 diagnosis. PCR can detect SARS-CoV-2 RNA from respiratory specimens (collected through a variety of means such as nasopharyngeal or oropharyngeal swabs). However, it is a time-consuming manual process, and known to often result in high false-negatives.
As an alternative to PCR, clinicians often utilize the radiography examination for COVID-19 screening, where chest radiography images (i.e., X-ray and/or computed tomography (CT)) are analyzed for visual indicators associated with COVID-19 viral infection. Several convolutional neural networks (CNN) have been published (mostly appearing as pre-prints) in recent weeks to tackle image-based COVID-19 diagnosis. These methods are either designed for CT or X-ray images. CT-based methods commonly adopt a 2-step process: (i) pulmonary region 2D/3D segmentation and (ii) lung classification into COVID19-infected, viral/bacterial pneumonia-infected, or healthy. Chest X-ray-based methods mostly use CNNs to perform this 3-way classification through adopting straight-forward off-the-shelf deep networks. These methods are mostly trained and validated on datasets of different origins and cohort size. Besides, most of the available COVID-19 sample sizes are smaller compared to the healthy sample sizes used in these studies. Further, many of the proposed COVID-19 classification, infection localization, and segmentation approaches are not open source.
In this project, we planned to explore and compile different publicly available CT and X-ray databases of COVID-19, non-COVID-19 respiratory diseases, and healthy cases. We also planned to study and implement different state-of-the-art methods as well as develope novel methods for COVID-19 classification, infection localization and segmentation.
Semi-supervised Deep Learning for COVID-19 Infection Segmentation
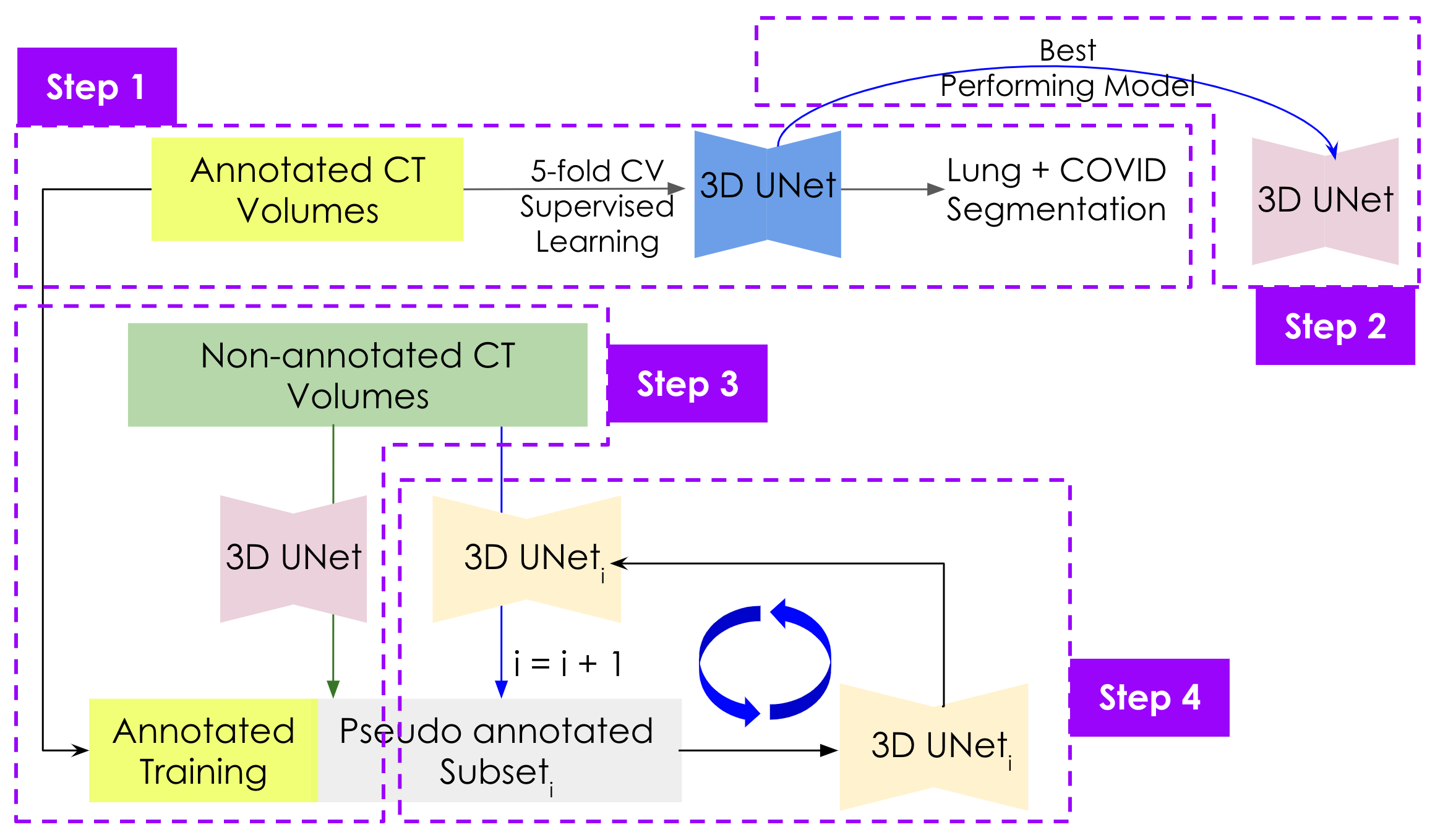
We used the 3D UNet with residual connections as the deep model. This method has 4 stepps: (1) At first, we use a 5-fold cross-validation to train the based model. (2) Then we select the best performing model for the later steps. (3) After that we used the best model to produce pseudo annotation for a small number of annotation-less data. (4) Finally, we added those newly annotated data to the expert-annotated datapool and finetune the model. We repeat steps 3 and 4 untill we finish iterating over the all the annotationless-data.
Data
We accessed a publicly available COVID-19 CT databases which contain pixel-level expert annotations of the COVID-19 infections. This dataset is called COVID-19 CT benchmark dataset (Ma et al. 2020) that contains 20 COVID-19 CT volumes from 20 patients with expert radiologists performed pixel-level annotation of COVID-19 infection in lung.
Results
The base model training run for 100 epochs/fold and the mean Dice metrics for five-fold cross-validations are 0.83, 0.88, 0.71, 0.68, and 0.74 respectively.

For Details
Please find the base 5-fold cross-validation code and trained model here.