Active Deep Learning from Noisy Teacher
Semi-Supervised COVID-19 Pneumonia Infection Segmentation in CT
Although reverse transcription polymerase chain reaction (RT-PCR) has been considered the gold standard for COVID-19 diagnosis, clinicians often utilize computed tomography (CT) scans to analyze the COVID-19 infection in lung. The automation of this infection analysis to accelerate clinical decision making typically requires pneumonia infection segmentation, where supervised deep learning has been showing promise. However, the difficulty of attaining pixel-level infection annotation for large volumetric CT images has recently steered the focus from developing supervised learning techniques to the investigation of semisupervised deep learning, so that a smaller expert-annotated dataset can be leveraged for infection segmentation in a larger dataset without annotation. However, semisupervised approaches often combine expert annotations and machine-generated annotation with equal weights in deep model training, though the later annotations are generally less reliable and potentially affect the model optimization negatively. To overcome this problem, we propose a semisupervised active learning approach that uses an example reweighting strategy, where machine-annotated training samples are weighted based on the similarity of their gradient directions to those of the expert-annotated data. We also adopt an active learning strategy by incorporating a query function in the reweighting algorithm to choose more trustworthy machine-annotated samples (i.e., noisy teacher) from the batch data. When validated on clinical benchmark data, our method showed better infection segmentation performance compared to state-of-the-arts.
Semi-Supervised Active Learning from Noisy Teacher
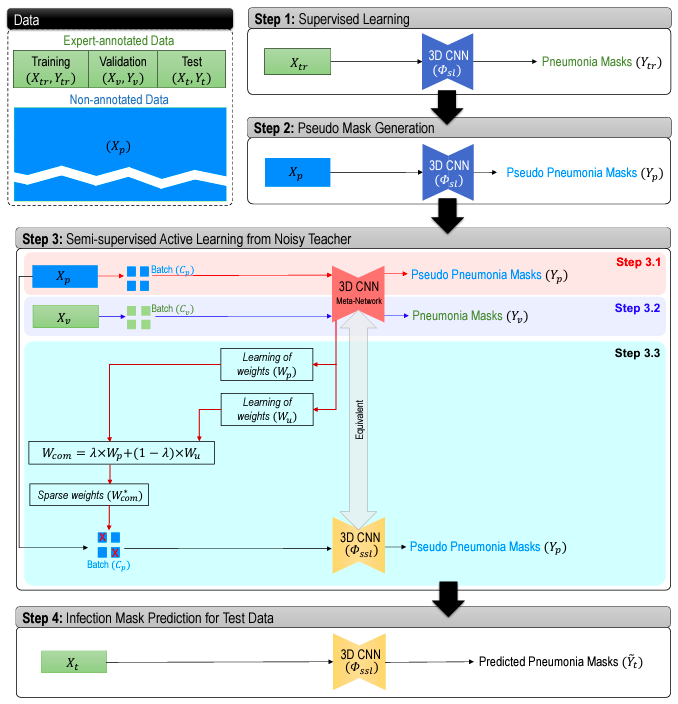
Brief Description of the Method We design the working pipeline of the proposed method using the following steps:
- Initially, we generate voxel-level annotations (pseudo annotation) using supervised deep models (step 1 in figure above) while considering the fact that these machine-generated annotations are less reliable (noisy teacher) than human expert annotations (step 2 in figure above).
- Then we generate a relative weight based on gradients per sample based on its “trustworthiness” during training. A sample weight is estimated from the similarity of the gradient directions between the annotation-free sample data and the expert-annotated validation data (step 3 in figure above).
- As the primary aim of an active learning algorithm is to identify and label only maximally-informative samples, gradient similarity-based sample weighting may lead to underestimation of a more diversely informative data sample. On the other hand, gradient magnitude with respect to parameters in the final CNN layer can be used as a measure of a model’s uncertainty. The higher magnitude of the gradient of the last layer, resulting from a higher loss of training, implies that the interrogated training sample contains newer information that the model has not yet seen. In our proposed approach, we adopt this gradient magnitude-based strategy and generate another set of sample weights based on their “informativeness” during training (step 3 in figure above).
- Afterwards, we generate an overall sample weight by combining the “trustworthiness” and “informativeness” sample weights.
- Finally, we use a query mechanism to choose more informative and trustworthy samples in a batch of annotation-free data by rectification (i.e., choosing more useful data in a batch) of the combined sample weight, and subsequently use these combined sample weights in the batch during the model optimization.
Data
We used three clinical datasets to validate our proposed method. We accessed two publicly available COVID-19 CT databases which contain pixel-level expert annotations of the COVID-19 infections. The first dataset is called COVID-19 CT benchmark dataset (Ma et al. 2020) that contains 20 COVID-19 CT volumes from 20 patients with expert radiologists performed pixel-level annotation of COVID-19 infection in lung. We also accessed the COVID-19 lung CT lesion segmentation challenge 2020’ database, which contains 199 unenhanced chest CT scans from 199 COVID-19 patients with ground truth pixel-level annotations of COVID-19 lesions in the lung. Finally, we accessed 1473 CT scans from 623 patients from ANONYMOUS hospital with all required ethics approvals in place. Among these scans, 567 are from COVID-19 and 906 are from CAP patients or healthy humans. However, none of these scans have pixel-level annotation.
Results
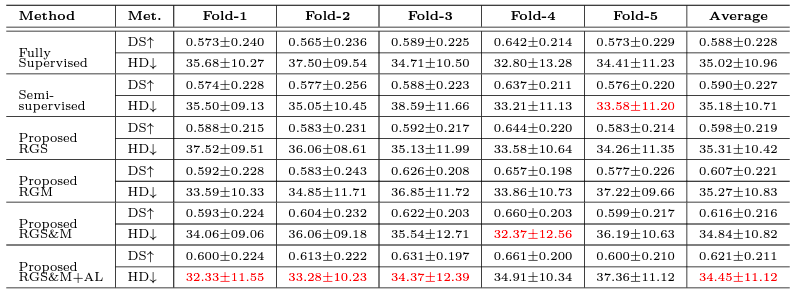
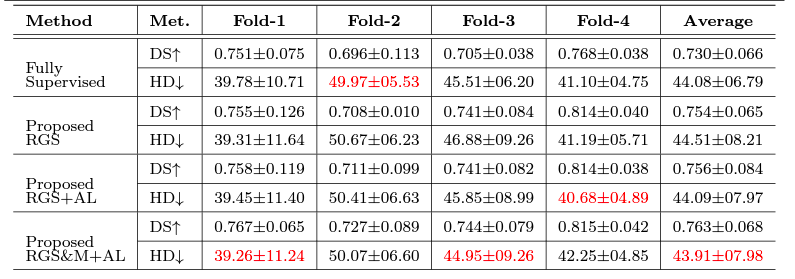
For Details
Please read our paper for details [1].